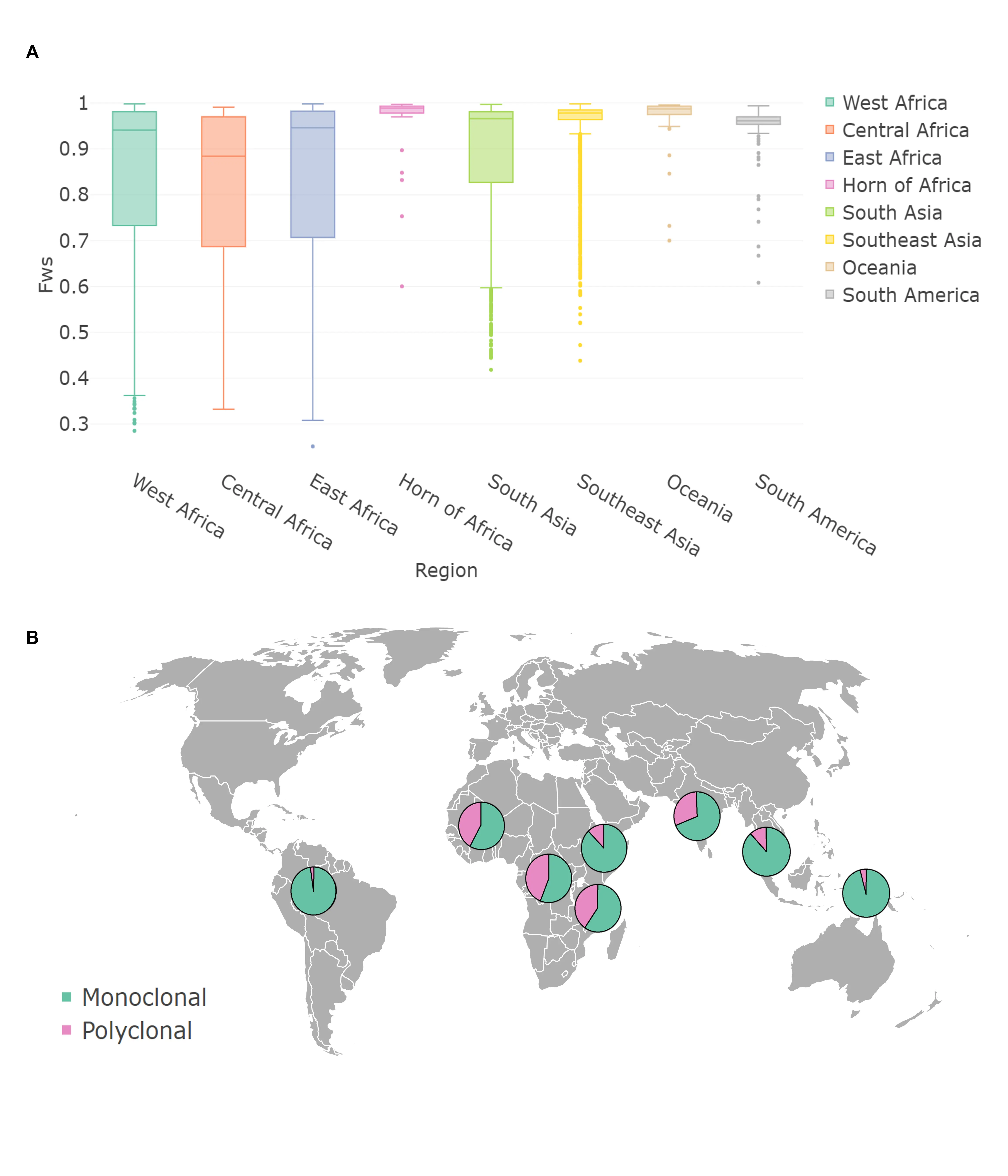
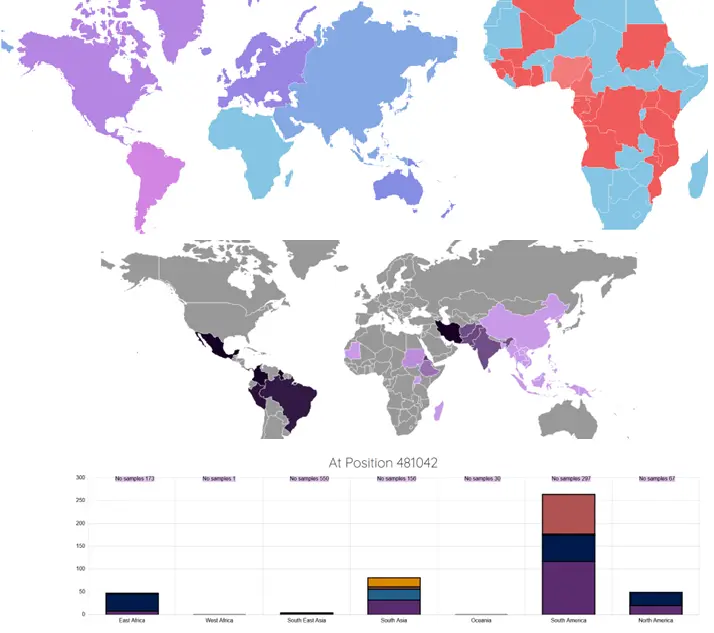
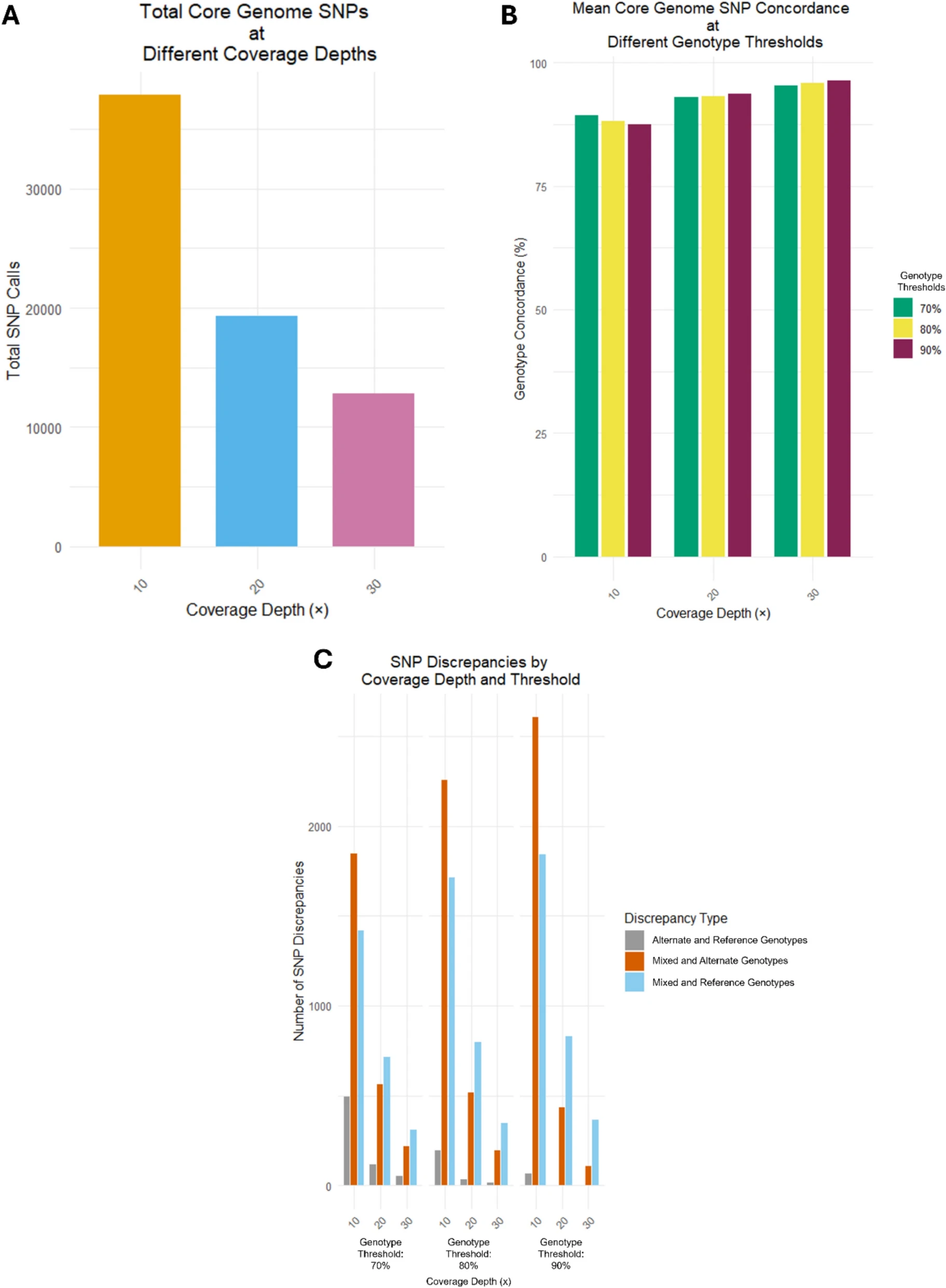
The malaria vector Anopheles stephensi has invaded and established itself in the Horn of Africa, and more recently Nigeria. The expansion of this vector poses a significant threat to malaria control and elimination efforts. Molecular methods are urgently needed to assist surveillance activities, including for the detection of insecticide resistance mutations. Work performed by Holly et al, presents a next-generation amplicon-sequencing approach, for high-throughput monitoring of insecticide resistance genes, species identification and characterization of genetic diversity in An. stephensi. Our work presents a reliable, cost-effective strategy using amplicon-sequencing to monitor known insecticide resistance mutations, with the potential to identify new genetic variants, to assist in the high-throughput surveillance of insecticide resistance in An. stephensi populations.
 Holly Acford-Palmer 5 April 2023
Holly Acford-Palmer 5 April 2023
During February and March, the Campino and Clark groups hosted 4 researchers from the Thailand Ministry of Public Health, who have been generating genomic and serological data as part of a (MRC-funded) COVID-19 vaccine break-out study. Thanks to Daniel Ward for mentoring the Thailand team
 Taane Clark 2 April 2023
Taane Clark 2 April 2023
In March, our team lunch event was at Siam Central. Our third lunch event of the year was a resounding success, with everyone who attended enjoying the great food and even better company. We were able to connect with colleagues we have not seen in person for a while, as well as get to know some of the newer team members better. The best part was that we all agreed to strictly no work talk, which allowed us to focus on building relationships and having fun.
 Joseph Thorpe 31 March 2023
Joseph Thorpe 31 March 2023